Here you can find detailed explanations on how you can use our web service to search for spatial residue patterns in proteins. You can easily click on the respective category to show the instructions. Brief answers to most usage-related questions are given in the FAQ section .
 | Before you can start the large-scale screening for a structural motif you will need: |
| download archive of predefined lists used by Fit3D |
To define a new structural motif you can use our extraction wizard that allows convenient and intuitive motif definition. You can either access a PDB structure by its PDB-ID or you can upload a structure in PDB format. The video below shows the extraction process step-by-step using a known PDB structure:
|
To submit a new job click "Submit job" in the left navigation menu. A motif file in PDB format is mandatory and must be provided by the user. Otherwise, the extraction wizard can be used to obtain a query structure. The target search space can be customized by uploading a list of PDB-IDs separated by line break. Alternatively, different pre-defined target lists, that were computed based on sequence or structure homology analysis, can be selected. The video below shows the submission process in detail:
|
Position-specific exchangeDefinitions (PSEs) for the motif search can be easily defined by using the advanced settings dialog. This can be useful for analysis of diverse protein families or mutagenesis experiments. PSEs are residue substitutions that are allowed during motif matching. As the name suggest, Fit3D handles position-specific residue substitutions, e.g. one can define a single residue in the query motif to be also matched against other residue types. The video below shows how to allow matching of different amino acid types:
| |
|
Note: The total number of position-specific exchangeDefinitions
(PSEs) is restricted to three.
|
To view the results of your job click "View results" in the left navigation menu. You can now select a corresponding job to show all detailed results. Watch the video below to see the whole process:
|
|
When viewing the results of your job, motif matches are ranked based on the lowest least-root-mean-square deviation (RMSD). Hints on how to understand and interpret the RMSD are given in the FAQ section. If you want to investigate a single match in detail, just click on "Show" or "Show in structure". Furthermore, you can directly access the corresponding Protein Data Bank (PDB) entry of the structure. Note that you can download all results or the RMSD distribution plot by clicking on the "Action" drop down menu on the toolbar at the very top of the result page. |
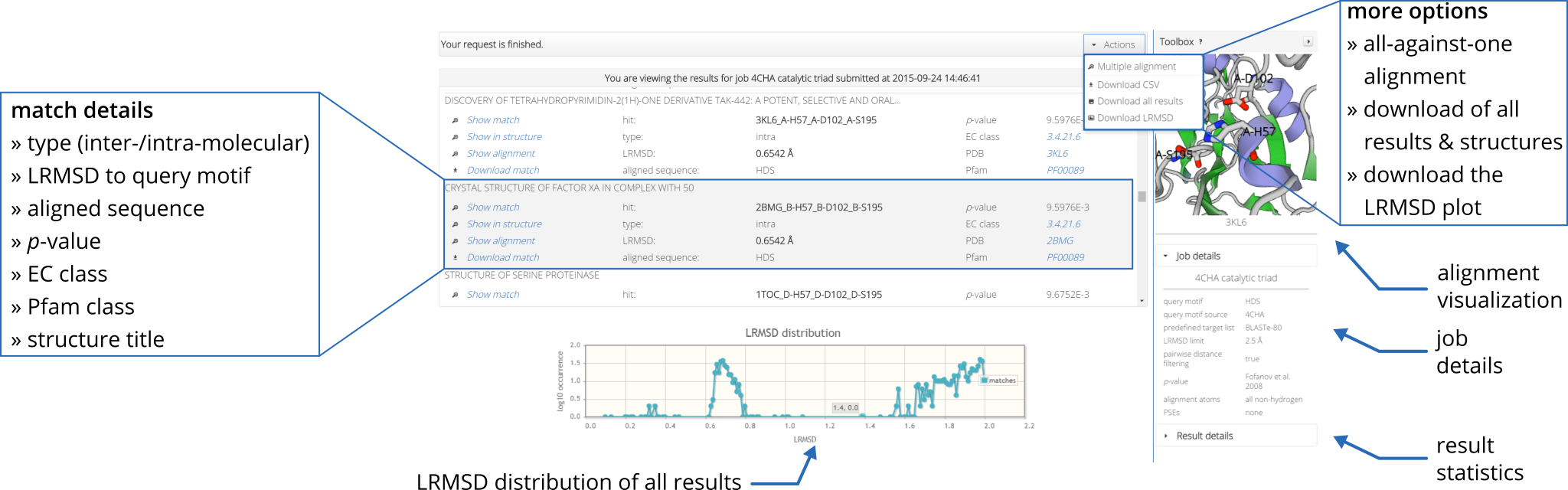
|
Beside geometric similarity matches are rated according their statistical significance. Every reported match to a query motif is labeled with a p-value such that the user can assess true positives intuitively. However, a significant p-value does not necessarily indicate a true positive match because the underlying models are based solely on the distribution of RMSD values. Hints on how to understand and interpret the p-value are given in the FAQ section. No biological features are represented by those models. To estimate statistical significance, Fit3D implements two statistical models: |
|